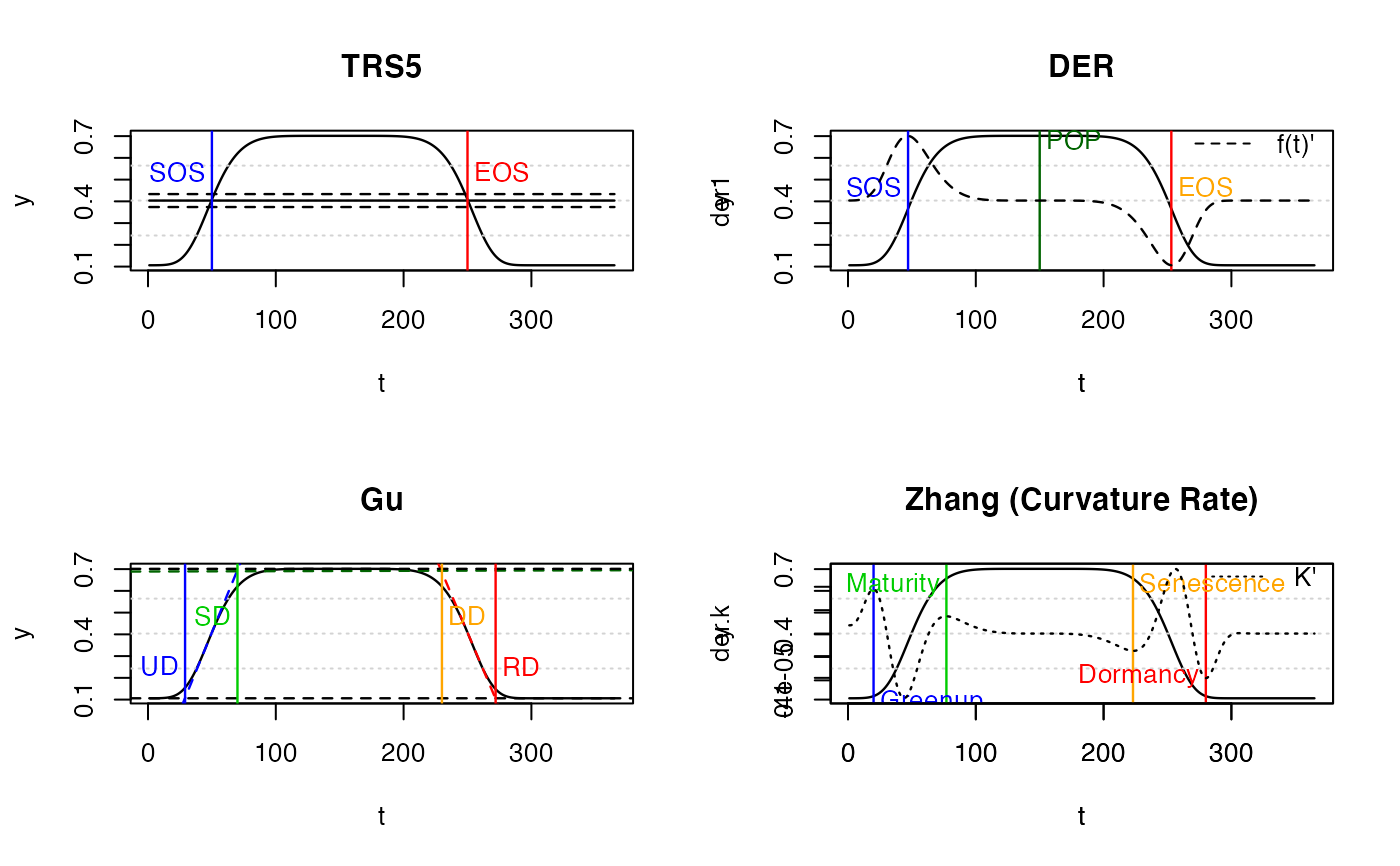
PhenoTrsThreshold methodPhenoDerivDerivative methodPhenoGuGu methodPhenoKlInflection method
PhenoTrs( fFIT, approach = c("White", "Trs"), trs = 0.5, asymmetric = TRUE, IsPlot = TRUE, ... ) PhenoDeriv( fFIT, analytical = TRUE, smoothed.spline = FALSE, IsPlot = TRUE, show.lgd = TRUE, ... ) PhenoGu(fFIT, analytical = TRUE, smoothed.spline = FALSE, IsPlot = TRUE, ...) PhenoKl( fFIT, analytical = TRUE, smoothed.spline = FALSE, IsPlot = TRUE, show.lgd = TRUE, ... )
Arguments
| fFIT |
|
|---|---|
| approach | to be used to calculate phenology metrics. 'White' (White et al. 1997) or 'Trs' for simple threshold. |
| trs | threshold to be used for approach "Trs", in (0, 1). |
| asymmetric | If true, background value in spring season and autumn season is regarded as different. |
| IsPlot | whether to plot? |
| ... | other parameters to PhenoPlot |
| analytical | If true, |
| smoothed.spline | Whether apply |
| show.lgd | whether show figure lelend? |
Examples
#> Error in library(phenofit): there is no package called ‘phenofit’# simulate vegetation time-series fFUN = doubleLog.Beck par = c( mn = 0.1 , mx = 0.7 , sos = 50 , rsp = 0.1 , eos = 250, rau = 0.1) t <- seq(1, 365, 8) tout <- seq(1, 365, 1) y <- fFUN(par, t) methods <- c("AG", "Beck", "Elmore", "Gu", "Zhang") # "Klos" too slow fFITs <- curvefit(y, t, tout, methods)#>#>#>#>#>#>#>#>#>#>#> sos eos #> 50 250PhenoDeriv(fFIT)#> sos pop eos #> 47 150 253PhenoGu(fFIT)#> UD SD DD RD #> 29 70 230 272PhenoKl(fFIT)#> Greenup Maturity Senescence Dormancy #> 20 77 223 280